Mila and Relation Therapeutics have been involved in a fruitful collaboration via the RECOVER project, funded in part by the Bill & Melinda Gates Foundation. The initial project, launched in 2020, aimed to discover a therapy against COVID-19 by identifying combinations of repurposed drugs which, together, can limit the entry of the virus inside human cells, and slow down its replication. To quickly discover a new therapy, we proposed to efficiently explore the space of drug combinations using an active learning and sequential model optimization pipeline. As a proof of concept, we applied our approach to the discovery of novel anti-cancer drug combinations due to the availability of public datasets.
Combining expertise in machine learning and experimental design was key to developing and implementing our new methodology. The machine learning pipeline predicts the response of cancer cells to combinations of drugs and proposes experiments for wet lab evaluation – ultimately leading to the discovery of novel combinations yielding synergies between drugs. Such an approach forms a powerful tool that could enable rapid responses to future public health crises.
Exploring the space of drug combinations
Drug combinations are an important therapeutic strategy for treating diseases that are subject to evolutionary dynamics, in particular cancers and infectious diseases [1, 2]. Conceptually, tumors and viruses can evolve or mutate over time and develop resistance to a single drug [3]. Combination therapies could play an important role in patient outcome by boosting drug efficacy and reducing toxicity, all while targeting multiple biological mechanisms simultaneously [4]. This reduces the chance of a mutation by a pathogen that would be simultaneously resistant to all the drugs in the combination. However, the space of drug combinations is still mostly unexplored due to the large combinatorial space of possible experiments — limiting the coverage of existing datasets and ultimately the quality of machine learning-based predictions. A cost-efficient solution involves the use of sequential model optimization (SMO) where one performs both informative experiments (“exploration”) and experiments that double-down on a promising hypothesis (“exploitation”) [5], the classical trade-off of reinforcement learning. Implementing such an approach could speed up the discovery of promising combinations for a given set of drugs while requiring substantially less experimentation.
In this work, we focus on pairwise drug combination synergy scores, and more precisely the Bliss synergy score, which indicates whether the combined effect of the two drugs is greater than expected outcome due only to their individual effects. Our objective in this work is the discovery of highly synergistic drug combinations which can activate the cell death program in human cancer cells cultivated in vitro. While SMO has already been applied to yeasts [6] and bacteria [7], focusing on human cells presented key technical challenges, such as batch-to-batch variability.
RECOVER: a sequential model optimization platform
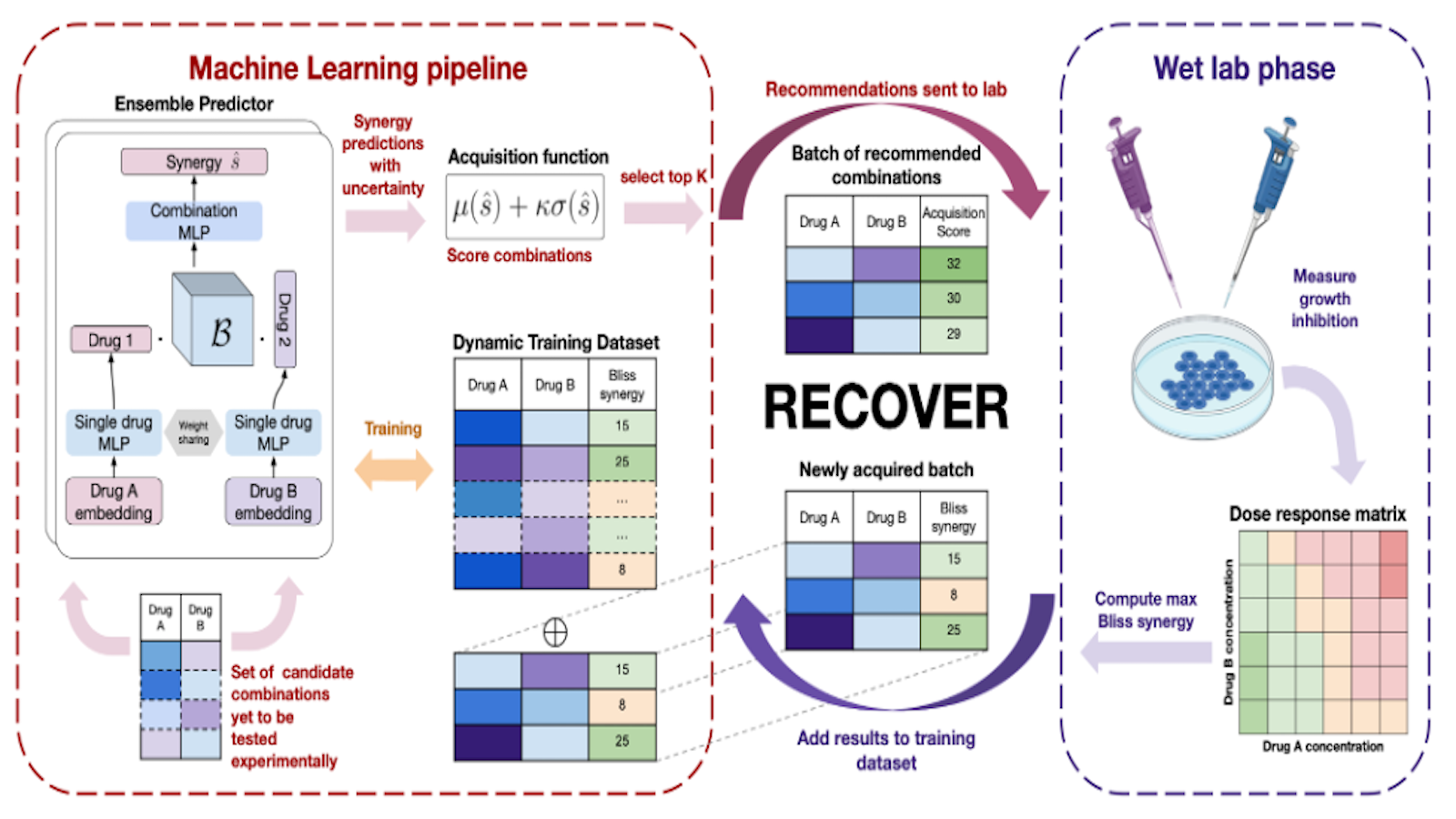
Our SMO setting (see Figure 1) generates recommendations of drug combinations that will be tested in vitro at regular intervals. At each step, our model is trained on all the data acquired up to that point. An acquisition function takes model predictions and uncertainties as input and computes a goodness score that favors candidate combinations which are expected to work well and for which an experiment would be informative (the model is uncertain about the outcome). The highest scoring combinations according to the acquisition function are then used to provide recommendations for in vitro testing. The results of the experiments are then added to the training data for the next round of experiments and the whole process repeats itself.
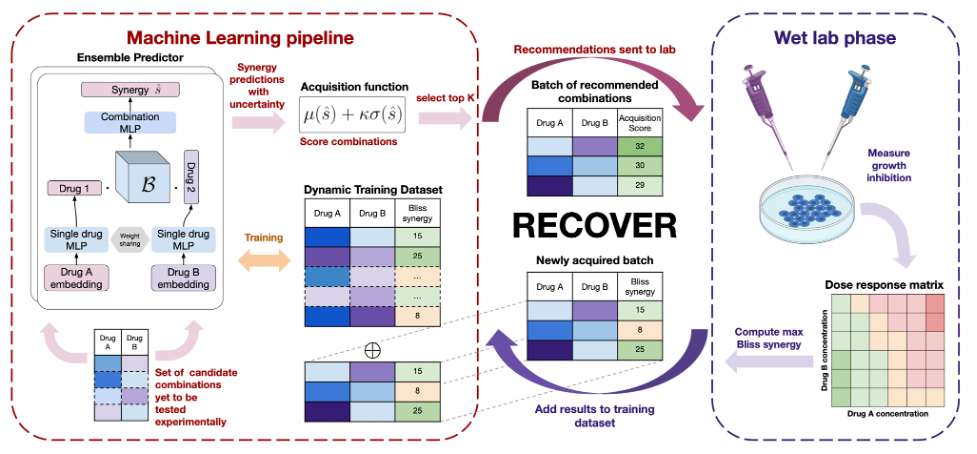
Figure 1. Overview of the RECOVER workflow integrating both a novel machine learning pipeline and iterated wet lab evaluation.
While we first experimented with advanced models relying on protein interaction databases and transcriptional signatures, we finally favored a simple model yielding very similar performance in order to ease the adoption of the pipeline by other practitioners. Pairs of drug feature vectors are fed into a deep neural network which is used for the prediction of synergy scores. These feature vectors include molecular fingerprints as well as one-hot encodings of the drugs.
The need for a sequential approach is confirmed by in silico evaluation
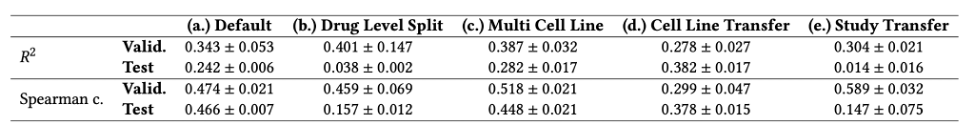
Our model was evaluated on different tasks, presented in Figure 2, to assess its generalization abilities. While performance can appear modest, we showed that they are reasonably close to the hypothetical maximum given the amount of experimental noise. As shown in Table 1, we noticed a significant drop in performance when trying to generalize to combinations of new drugs. However, performance is still above the level of random selection. Second, we noticed that leveraging several distinct populations of cancer cells (called cell lines) improves performance on the cancer cells of interest, in terms of R2. Finally, we noticed that the model generalizes poorly to a new experimental setting.
For prospective in vitro experiments, this analysis suggests a sequential approach is required to adapt our model to a new laboratory and then to novel chemical compounds. This makes it possible to find a good combination much faster than if we had followed the traditional machine learning practice of first collecting data, then training a model, then using the trained model to screen a large number of candidate combinations.
Figure 2. Overview of the different tasks on which RECOVER has been evaluated. Each task corresponds to a different way to split the training, validation and test sets, and aims at evaluating a specific generalization ability of the model.
Table 1. Performance of RECOVER for the different tasks, as detailed in Figure 2. Standard deviation computed over 3 seeds.
In silico testing of RECOVER rapidly predicts and rediscovers synergistic drug combinations
Our in silico experiments tried to mirror as closely as possible the setting of the in vitro experiments. We compare several search strategies by the rate at which they unblind the set of top 1% of synergistic combinations in the dataset within each synthetic experimental round (or iteration). As shown in Figure 3, Greedy and Upper Confidence Bound (UCB) acquisition functions, two Model-based searches, perform on par with each other and outperform Random Search by a large margin.
In other in silico experiments (not matching our in vitro setting), UCB outperformed the Greedy acquisition function, see Figure 5 (inset). This demonstrates that taking uncertainty into account to guide experiments can increase the performance of the pipeline over a naive Greedy acquisition strategy in some cases.
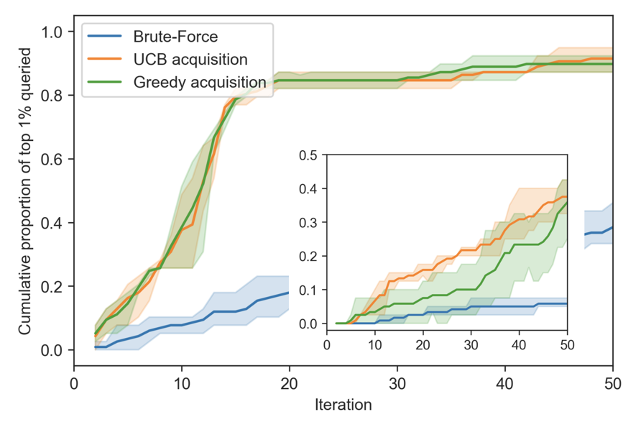
Figure 3. Cumulative proportion of the top 1% synergistic combinations that have been rediscovered by RECOVER. (inset) UCB can outperform Greedy acquisition in certain configurations. Standard deviation computed over 3 seeds.
Prospective use of RECOVER discovers and rediscovers novel synergistic drug combinations
From the in silico results, we then tested RECOVER prospectively with the Lairson lab at The Scripps Research Institute: we performed three rounds of RECOVER-informed wet lab experiments and observed sequential improvements in performance. While the first two rounds were focused on model calibration and ensuring a reasonable proportion of combination space was observed, the final round of nominated drug combinations reflected a real-world use case with minimal human oversight.
The search space consisted of 2,916 drug combinations (see Figure 4A). In each round, the synergies of 30 combinations were evaluated. In Figure 4B, we plot the cumulative density function of each experimental round. We note that the distribution starts developing a heavier tail towards high maximum Bliss synergy scores. This emergent heavy tail appears significant when comparing the distribution in the SMO Search round to the background distribution of synergy scores from the public dataset (Kolmogorov–Smirnov test, p < 0.025). Finally, the highest max Bliss synergy score observed increases from one round to the next.
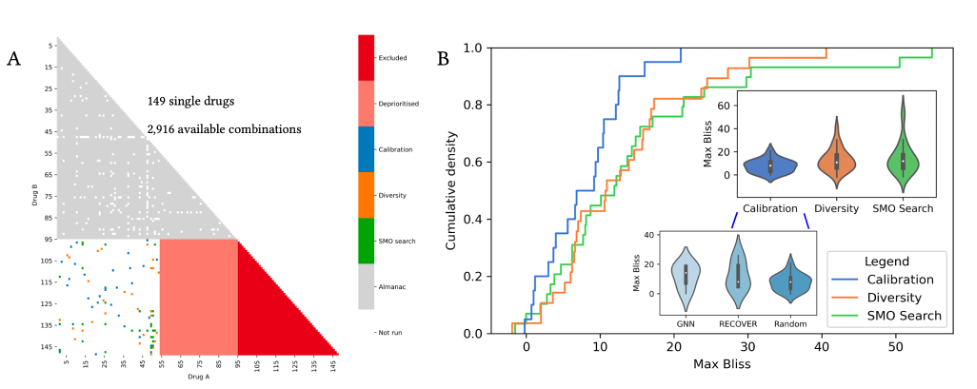
Figure 4. Prospective use of RECOVER for in vitro evaluation. (A.) Diagram representing drug combinations used during pretraining, in three subsequent experiment rounds, and excluded from the analysis. Drug combinations that were not available for pretraining or were not selected for experiments are represented in white. (B.) Cumulative density plot of max Bliss synergy score for each experimental round.
Conclusion
We have presented the SMO pipeline RECOVER for drug combination selection. We showcase a general methodology, consisting of careful analysis of the properties of the machine learning pipeline – such as its out-of-distribution generalization capacities – to help us design some aspects of the in vitro experiments, and eventually ensure a smooth and successful interaction between the machine learning pipeline and the wet lab.
After only three rounds of ML-guided in vitro experimentation, we found that the set of combinations queried by our model was enriched for highly synergistic combinations. Remarkably, we rediscovered a synergistic drug combination that was later confirmed to be under study within clinical trials [8].
This blog post is based on our recent preprint: https://arxiv.org/pdf/2202.04202.pdf
Code: https://github.com/RECOVERcoalition/Recover
Acknowledgments
This research is funded in part by the Bill & Melinda Gates Foundation. The findings and conclusions contained within are those of the authors and do not necessarily reflect positions or policies of the Bill & Melinda Gates Foundation. The authors would also like to thank Isabelle Lacroix for useful support.
Paper Authors
- Paul Bertin, Mila
- Jarrid Rector-Brooks, Mila
- Deepak Sharma, Mila
- Thomas Gaudelet, Relation Therapeutics
- Andrew Anighoro, Relation Therapeutics
- Torsten Gross, Relation Therapeutics
- Francisco Martínez-Peña, Department of Chemistry, The Scripps Research Institute
- Eileen Tang, Department de Chemistry, The Scripps Research Institute
- Suraj M S, Relation Therapeutics
- Christian Regep, Relation Therapeutics
- Jeremy Hayter, Relation Therapeutics
- Maksym Korablyov, Mila
- Nicholas Valiante, Glyde Bio, Inc.
- Almer van der Sloot, Institute for Research in Immunology and Cancer (IRIC)
- Mike Tyers, Institute for Research in Immunology and Cancer (IRIC)
- Charles Roberts, Relation Therapeutics
- Michael Bronstein, Department of Computer Science, Université d’Oxford, Twitter
- Luke Lairson, Department of Chemistry, The Scripps Research Institute
- Jake Taylor-King, Relation Therapeutics
- Yoshua Bengio, Mila
References
[1] Adrian Schmid, Aline Wolfensberger, Johannes Nemeth, Peter W Schreiber, Hugo Sax, and Stefan P Kuster. Monotherapy versus combination therapy for multidrug-resistant gram-negative infections: Systematic review and meta-analysis. Scientific reports, 9(1):1–11, 2019.
[2] Reza Bayat Mokhtari,Tina S Homayouni, Narges Baluch, Evgeniya Morgatskaya, Sushil Kumar, Bikul Das, and Herman Yeger. Combination therapy in combating cancer. Oncotarget, 8(23):38022, 2017.
[3] João Delou, Alana SO Souza, Leonel Souza, and Helena L Borges. Highlights in resistance mechanism pathways for combination therapy. Cells, 8(9):1013, 2019.
[4] Bissan Al-Lazikani, Udai Banerji, and Paul Workman. Combinatorial drug therapy for cancer in the post-genomic era. Nature Biotechnology, 30(7):679-692, July 2012.
[5] Yuriy Sverchkov and Mark Craven. A review of active learning approaches to experimental design for uncovering biological networks. PLoS computational biology, 13(6):e1005466, 2017.
[6] Ross D. King, Kenneth E. Whelan, Ffion M. Jones, Philip G. K. Reiser, Christopher H. Bryant, Stephen H. Muggleton, Douglas B. Kell, and Stephen G. Oliver. Functional genomic hypothesis generation and experimentation by a robot scientist. Nature, 427(6971):247–252, Jan 2004.
[7] Pablo Carbonell, Adrian J. Jervis, Christopher J. Robinson, Cunyu Yan, Mark Dunstan, Neil Swainston, Maria Vinaixa, Katherine A. Hollywood, Andrew Currin, Nicholas J. W. Rattray, Sandra Taylor, Reynard Spiess, Rehana Sung, Alan R. Williams, Donal Fellows, Natalie J. Stanford, Paul Mulherin, Rosalind Le Feuvre, Perdita Barran, Royston Goodacre, Nicholas J. Turner, Carole Goble, George Guoqiang Chen, Douglas B. Kell, Jason Micklefield, Rainer Breitling, Eriko Takano, Jean-Loup Faulon, and Nigel S. Scrutton. An automated design-build-test-learn pipeline for enhanced microbial production of fine chemicals. Communications Biology, 1(1):66, Dec 2018.
[8] Hiral A Shah, James H Fischer, Neeta K Venepalli, Oana C Danciu, Sonia Christian, Meredith J Russell, Li C Liu, James P Zacny, and Arkadiusz Z Dudek. Phase i study of aurora a kinase inhibitor alisertib (mln8237) in combination with selective vegfr inhibitor pazopanib for therapy of advanced solid tumors. American journal of clinical oncology, 42(5):413–420, 2019.